Computational and Systems Biology
Perhaps the most active interdisciplinary research arena is the intersection of the life sciences with informatics. Understanding how the cell regulates collections of genes in evolutionary interaction with the environment, other organisms, drugs, and even society is increasingly done by merging biology with informatics and other areas of computational science, including artificial intelligence. Methods and theory from information retrieval, text mining, knowledge discovery, machine learning, computational modeling, complex networks and systems theory, and data science, present us with the opportunity to make new discoveries in biology, medicine, and public health. In our CASCI group we are working on various projects which use such methods to aid biomedical discovery in both health-related and theoretical questions.

Biomedical Literature and Social Media Mining
Our approach to literature mining is based on network science, complex systems, machine learning, and bio-inspired methods, which we have applied to text classification, relational inference and annotation of protein-protein and drug-drug interactions, pharmacokinetics numerical data, protein sequence family and structure prediction, functional annotation of transcription data, enzyme annotation, and so on. We have applied our methods to the published scientific literature, social media, electronic health records, bioinformatics databases, etc.
MoreModeling of biochemical regulation dynamics
The paradigmatic example of a complex system is the web of biochemical interactions that make up life. We still know very little about the organization of life as a dynamical, interacting network of genes, proteins and biochemical reactions. We are focused on developing network and dynamical systems methodologies and informatics tools to study control, modularity, robustness, evolvability, and collective computation in automata networks used to model gene regulation and biochemical signaling. We are also using such methods to study the interplay between network structure and dynamics in the brain.
More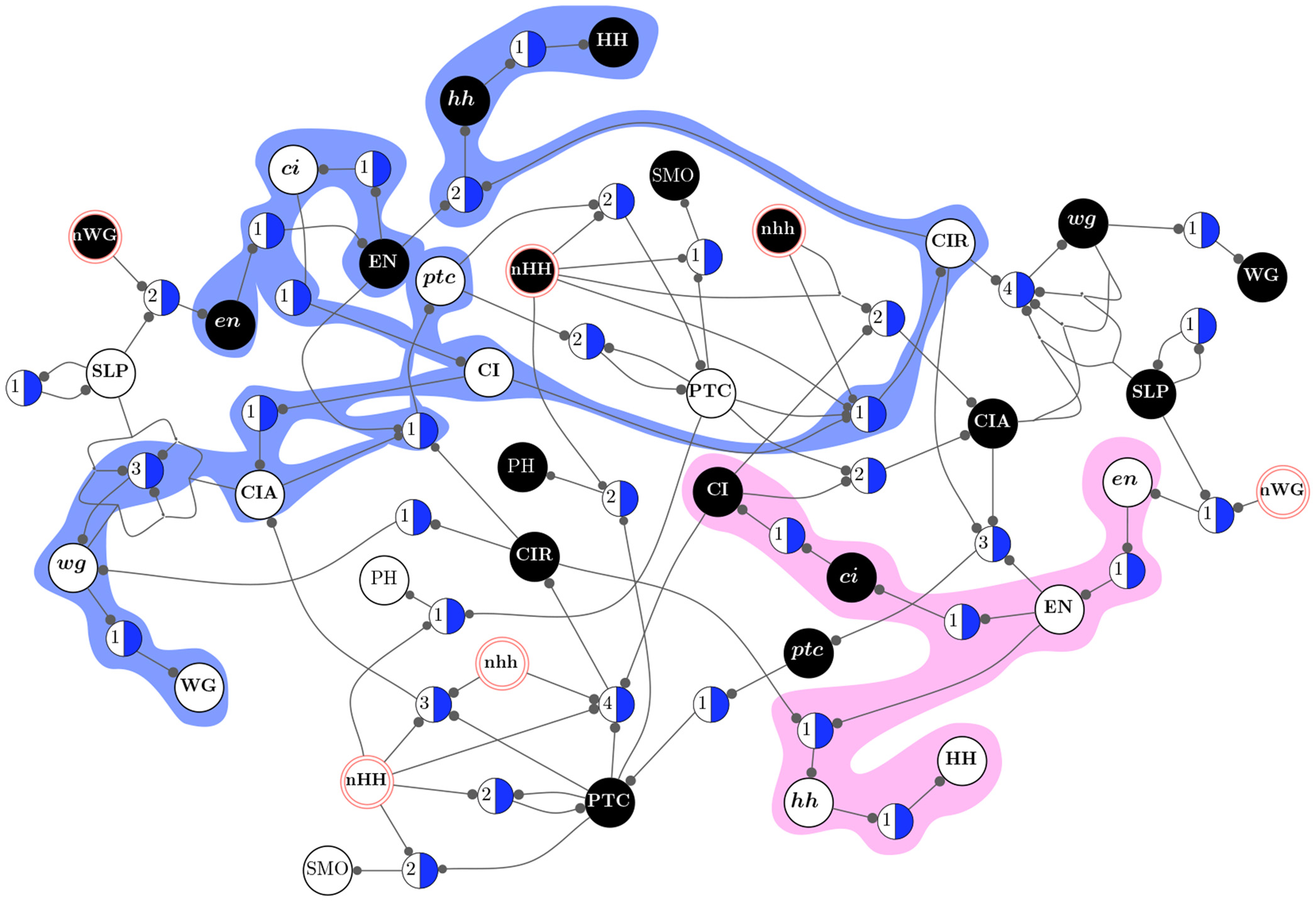
Multivariate Time-Series Analysis and Network Inference
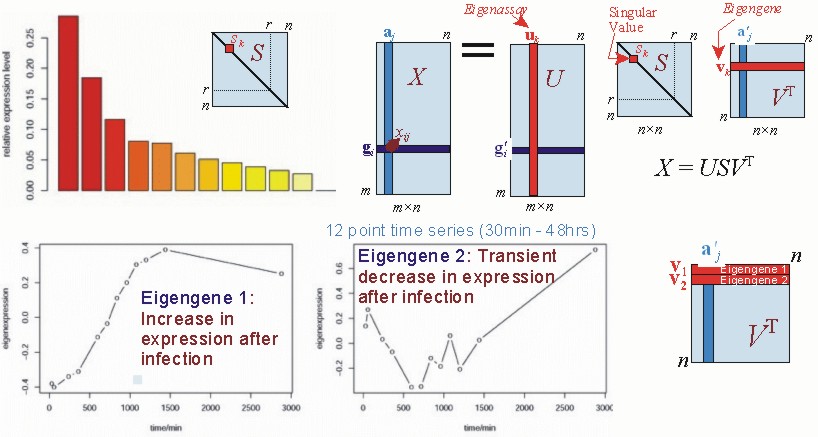
In the age of biomedical data-science, it is essential to develop methods to infer time-varying data associations such as pairwise variable interactions and subsets of variables that mostly interact with one another (data clustering, network modularity). Our contribution to the problem of inference on networks and multivariate dynamics has been in the area of spectral methods, statistical inference, and information theory, which has been used to uncover interactions and multiscale modularity in various domains, such as gene regulation, transcriptomics and brain activity time-series data. In particular, we have worked in clustering methods for various types of genomic data which allow multiple membership of genes in clusters. With various collaborators, we became very interested in using spectral analysis, such as Singular Value Decomposition (SVD), as an automated method for Functional Genomics. These methods have been extended to uncovering modularity in networks, allowing the identification of overlapping functional clusters that occur in various scales of complex networks.
MoreAgent-Based Modeling of Evolutionary Systems
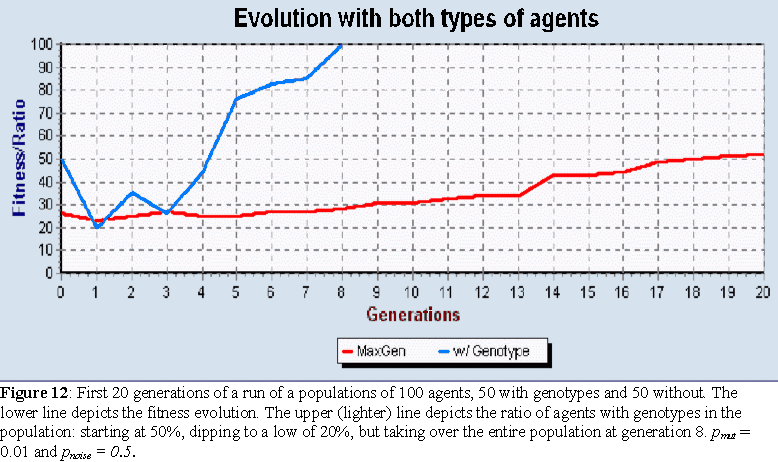
We have contributed to theoretical biology by developing agent-based models of systems whose evolutionary role or adaptive capabilities are not well understood. For instance, we provided the first computational model that demonstrates the evolutionary potential of RNA Editing and a model that proves the ability of T-Cell cross-regulation to classify self from non-self in the presence of populations of changing pathogens. We have also developed bio-inspired methods for spam detection and (biomedical) text classification from that model. More generally, we have contributed to the study of the interplay between self-organization and natural selection by introducing the concept of selected self-organization and developing evolutionary algorithms and agent-based models to study it. We have been working on various other applications of the agent based modeling framework, such as evolving cellular automata and adaptive agents for recommendation systems.
MoreBiosemiotics: interplay between self-organization and selection.
As Howard Pattee observed, while processes of a seemingly informational and indeed linguistic nature are fundamental to evolution in biology, computers which are based on the purely syntactic aspects of language are very non-adaptive. Therefore, we are interested in the linguistic/symbolic aspects of the living organization (the gene as a carrier of information, and DNA as memory) which play a large role in the seemingly open-ended evolution defined by natural selection. We study the interplay between self-organization and natural selection, focusing on the concept of selected self-organization. We are particularly interested in the problem of how information, symbols, representations and the like can arise from a purely dynamical system of many components.
More